This is the first part of the Machine Learning series. For your convenience you can find other parts using the links below (or by guessing the address):
Part 1 — Linear regression in MXNet
Part 2 — Linear regression in SQL
Part 3 — Linear regression in SQL revisited
Part 4 — Linear regression in T-SQL
Part 5 — Linear regression
Part 6 — Matrix multiplication in SQL
Part 7 — Forward propagation in neural net in SQL
Part 8 — Backpropagation in neural net in SQL
In this series I assume you do know basics of machine learning. I will provide some source code for different use cases but no extensive explanation. Let’s go.
Today we will take a look at linear regression in MXNet. We will predict sepal length in well know iris dataset.
I assume you have the dataset uploaded to s3. Let’s go with loading the dataset:
|
1 2 3 4 5 6 7 8 9 |
from mxnet import nd, autograd import pandas as pd import matplotlib.pyplot as plt from sklearn.model_selection import train_test_split local_file="/tmp/Iris.csv" df = pd.read_csv("/blabla/" + local_file, delimiter=',', header = 0) print df.shape |
We can see some records with print df.head(3) or check different iris categories with df.iris.unique().
We have one target variable and four features. Let’s create two adttional:
|
1 2 3 4 |
df['i_setosa'] = 0 df.loc[(df['iris']=='setosa'), 'i_setosa']= 1 df['i_versicolor'] = 0 df.loc[(df['iris']=='versicolor'), 'i_versicolor']= 1 |
Two features similar to one hot encoding of categorical feature.
Time to prepare training and test datasets with: df_train, df_test = train_test_split( df, test_size=0.3, random_state=1)
Let’s get down to training. We start with defining the training variables and the target one:
|
1 2 3 4 5 |
independent_var = ['sepal_width','petal_length','petal_width','i_setosa','i_versicolor'] y_train = nd.array(df_train['sepal_length']) X_train = nd.array(df_train[independent_var]) y_test = nd.array(df_test['sepal_length']) X_test = nd.array(df_test[independent_var]) |
Let’s prepare class representing data instance:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
class data: def __init__(self,X,y): self.X = nd.array(X) self.y = nd.array(y) cols = X.shape[1] self.initialize_parameter(cols) def initialize_parameter(self,cols): self.w = nd.random.normal(shape = [cols, 1]) self.b = nd.random.normal(shape = 1) self.params = [self.w, self.b] for x in self.params: x.attach_grad() |
We initialize parameters and attach gradient calculation. This is a very nice feature, we don’t need to take care of derivatives, everything is taken care for us.
Let’s now carry on with a single step for gradient:
|
1 2 3 4 5 6 7 |
class optimizer: def __init__(self): pass def GD(self,data_instance,lr): for x in data_instance.params: x[:] = x - x.grad * lr |
We just subtract gradient multiplied by learning rate. Also, we use x[:] instead of x to avoid reinitializing the gradient. If we go with the latter, we will see the following error:
|
1 |
Check failed: !AGInfo::IsNone(*i) Cannot differentiate node because it is not in a computational graph. You need to set is_recording to true or use autograd.record() to save computational graphs for backward. If you want to differentiate the same graph twice, you need to pass retain_graph=True to backward. |
Now, let’s train our model:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 |
def main(): # Modeling parameters learning_rate = 1e-2 num_iters = 100 data_instance = data(X_train,y_train) opt = optimizer() gd = optimizer.GD loss_sequence = [] for iteration in range(num_iters): with autograd.record(): loss = nd.mean((nd.dot(X_train, data_instance.w) + data_instance.b - y_train)**2) loss.backward() gd(opt, data_instance, learning_rate) print ("iteration %s, Mean loss: %s" % (iteration,loss)) loss_sequence.append(loss.asscalar()) plt.figure(num=None,figsize=(8, 6)) plt.plot(loss_sequence) plt.xlabel('iteration',fontsize=14) plt.ylabel('Mean loss',fontsize=14) |
We should get the following:
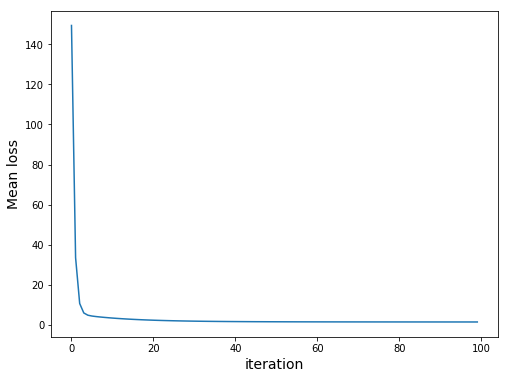
Note that our loss uses sum. However, due to overflow problems we would get the following:
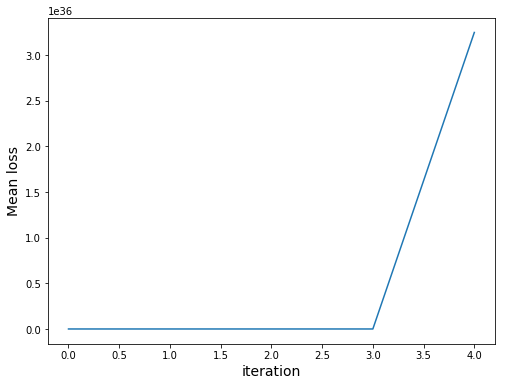
Finally, let’s check the performance of trained model:
|
1 2 |
MSE = nd.mean(((nd.dot(X_test, data_instance.w) + data_instance.b) - y_test)**2) print ("Mean Squared Error on Test Set: %s" % (MSE)) |
Done.
Summary
We can see that linear regression is pretty concise and easy. However, this uses Python and Spark which me might want to avoid. In next parts we will take a look at different solutions.